Abstract
Biosynthetic gene clusters (BGCs) are genomic loci encoding the biosynthetic pathway for producing specialised metabolites
with a broad spectrum of bioactivities. Many methods have been developed for genome mining of BGCs, such as GECCO.
However, in many cases the cognate metabolite remains unknown, and experimental characterisation remains difficult.
CHAMOIS is a machine learning-based tool for predicting chemical properties of secondary metabolites from protein domains annotated in the input BGCs.
CHAMOIS infers 539 chemical properties from the ChemOnt ontology using logistic regression. It accurately predicts 111 such properties (AUPRC > 0.5)
in cross-validation against known instances. Although CHAMOIS is not explicitly trained on biosynthetic knowledge, many of the inferred
links between protein domains and metabolite properties are consistent with scientific literature, others suggest new biochemical functions of
uncharacterized biosynthetic domains. Finally, CHAMOIS can pinpoint which BGC within a given genome produces a pre-specified metabolite
(correct BGC in 72% of cases ranked among the top 5), which holds great potential for prioritising experimental BGC characterisation and discovery
of novel biosynthetic enzymes.
The CHAMOIS software is implemented in Python,
supports all versions from Python 3.7 and is provided under
the GNU General Public License v3.0 or later.
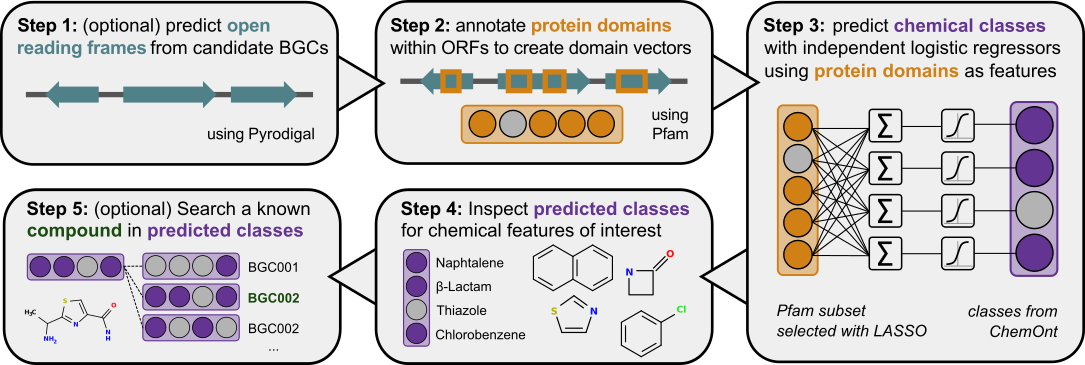
Graphical depiction of the chemical hierarchy inference approach implemented in CHAMOIS. Briefly, CHAMOIS
identifies open reading frames (ORFs) in a given BGC sequence (Step 1). Then, protein domains
are annotated in the resulting ORFs using profile hidden Markov models (pHMMs; Step 2). The resulting domain
vector for the whole BGC serves as a feature for a logistic regression classifier for each class of the
ChemOnt ontology (Step 3). Predicted classes allow filtering for BGCs with particularly relevant chemical classes (Step 4).
Finally, the fingerprint of class predictions can be used to find BGCs most similar to a particular compound (Step 5).