Abstract
The human microbiome has been shown to exhibit distinct differences in many host diseases is increasingly mined for diagnostic and therapeutic biomarkers using machine learning (ML). However, metagenomics-specific and user-friendly software is scarce, and overoptimistic evaluation and limited cross-study generalisation are prevailing issues due to common pitfalls in model training and evaluation. Members of the Zeller lab have thus we developed SIAMCAT, a versatile R toolbox for ML-based comparisons of microbiomes in case/control settings. SIAMCAT provides complete workflows supporting data preprocessing, statistical association testing, statistical modelling and visualizations faciliating evaluation and interpretation of these models and includes important checks and balances to avoid common pitfalls. Additionally it provides functionality for analysis and post hoc correction of confounders as well as for cross-study comparison and meta-analysis.
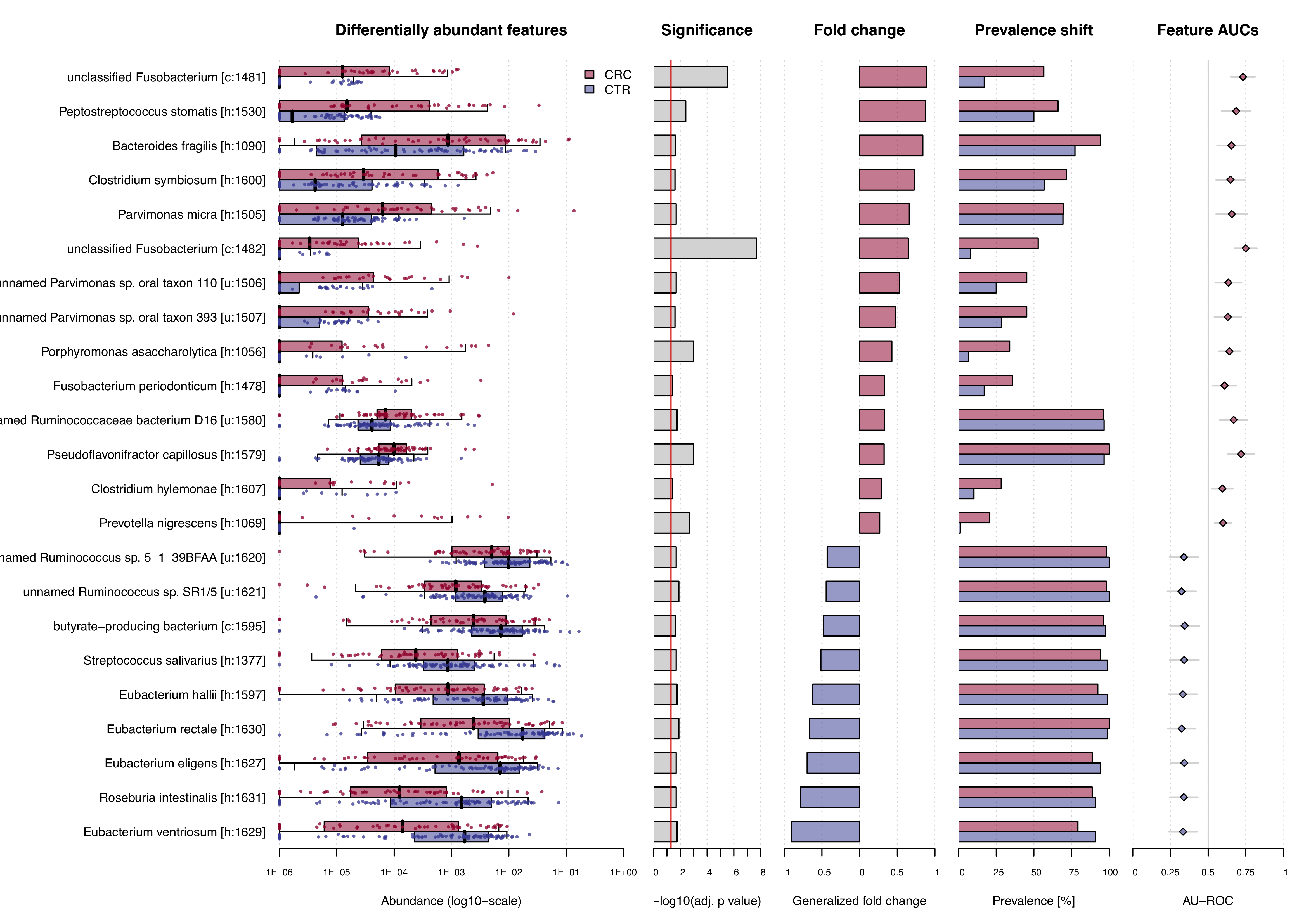
SIAMCAT is distributed under the GPL-3 license.
SIAMCAT has been used in the following publications:
- Cross-cohort gut microbiome associations with immune checkpoint inhibitor response in advanced melanoma
- A faecal microbiota signature with high specificity for pancreatic cancer
- MGnify: the microbiome sequence data analysis resource in 2023
- Candida expansion in the gut of lung cancer patients associates with an ecological signature that supports growth under dysbiotic conditions
- Impact of international travel and diarrhea on gut microbiome and resistome dynamics