Abstract
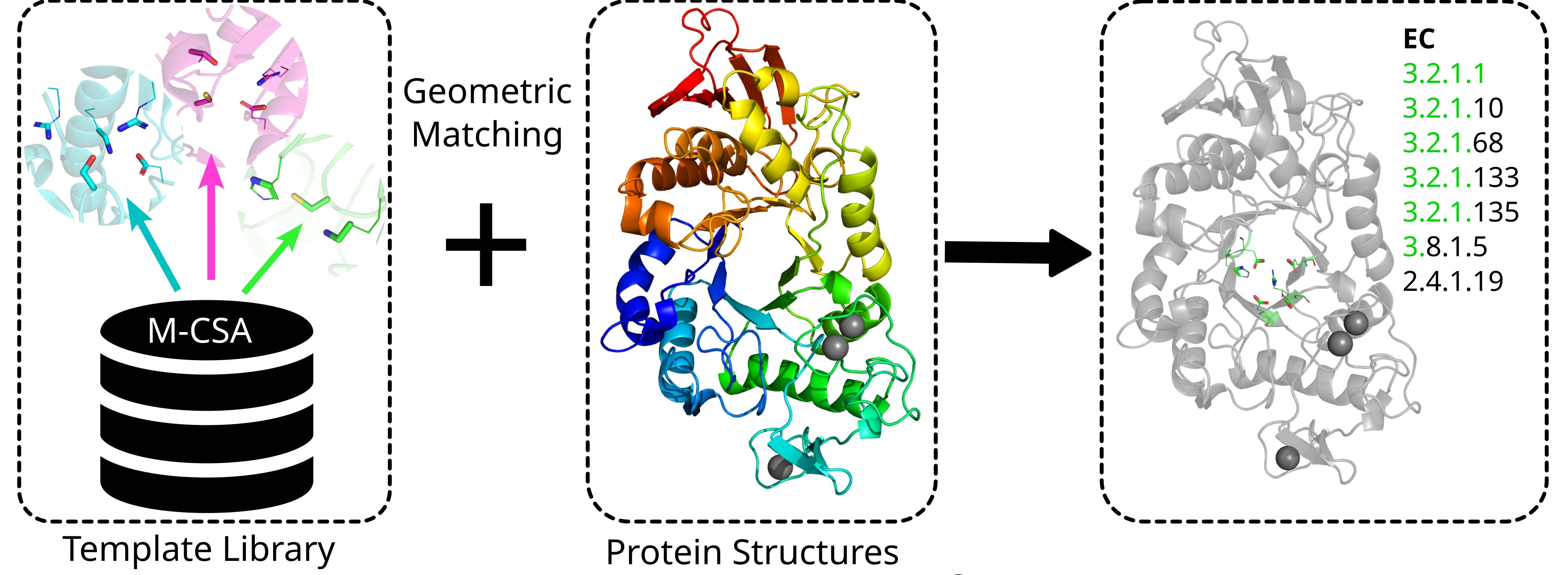
Enzyme Motif Miner, EnzyMM for short, is a computational tool to identify catalytic
sites in protein structures. It searches protein structures provided by the user
against a database of templates. EnzyMM ships with a library of catalytic templates
derived from the Mechanism and Catalytic Site Atlas (M-CSA) but you can also
generate your own. These templates represent consensus arrangements of catalytic sites
found in active sites of experimental protein structures.
By comparing the geometric orientation of amino acids, rather than relying on sequence
similarity, our approach is very fast while remaining interpretable.
We can uncover distant relationships between enzymes, suggesting how functions
may have evolved. Keeping pace with structure prediction, this method is well
suited for annotating catalytic sites across entire proteomes.
Enzyme Motif Miner is implemented in Python, supports all versions from Python 3.7 and is provided under an MIT Licence.
For the actual geometric matching EnzyMM relies on PyJess - a Cython wrapper of Jess.
If you just want to try EnzyMM we provide a webserver at https://www.ebi.ac.uk/thornton-srv/m-csa/enzymm .